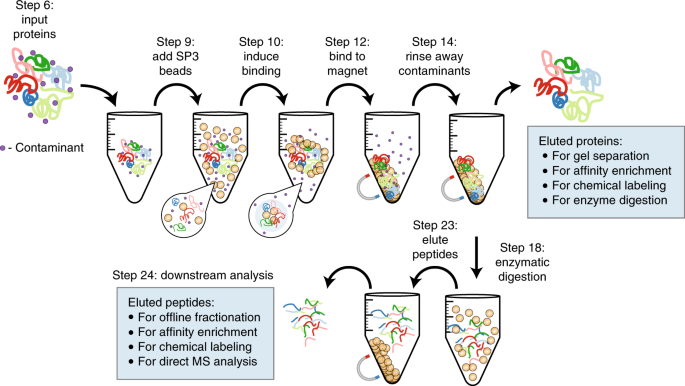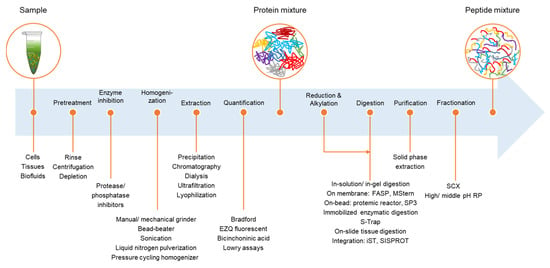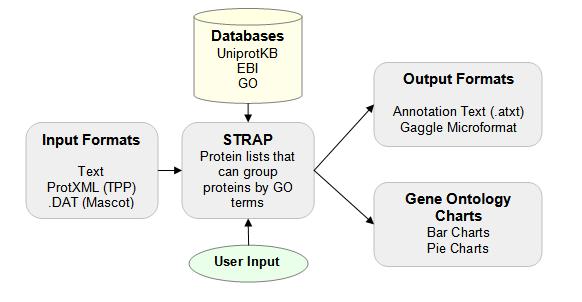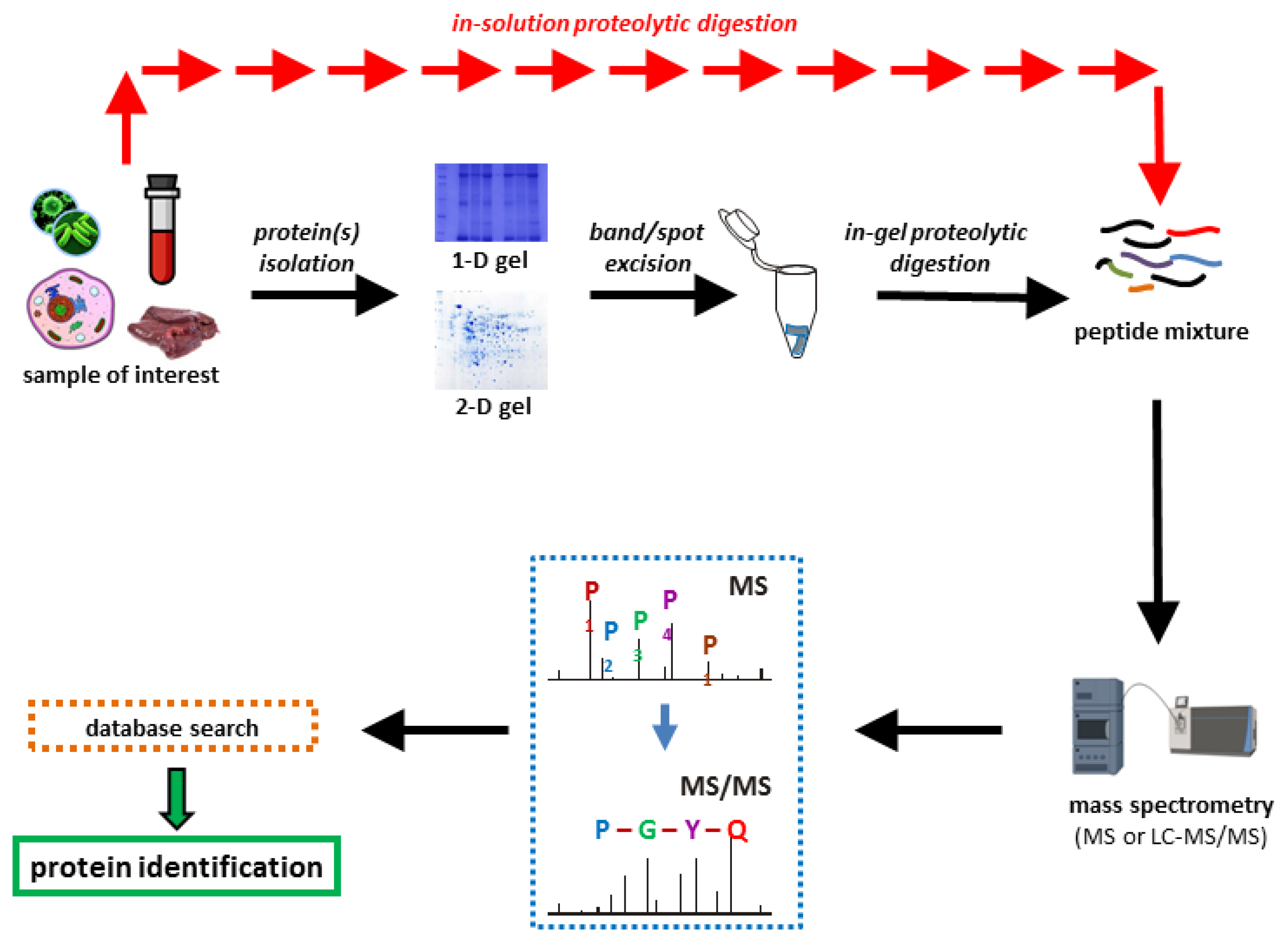
Proteomes | Free Full-Text | A Critical Review of Bottom-Up Proteomics: The Good, the Bad, and the Future of This Field

Comparison of In-Solution, FASP, and S-Trap Based Digestion Methods for Bottom-Up Proteomic Studies | Journal of Proteome Research

Suspension trapping (STrap) sample preparation method for bottom‐up proteomics analysis - Zougman - 2014 - PROTEOMICS - Wiley Online Library

Self-Assembled STrap for Global Proteomics and Salivary Biomarker Discovery | Journal of Proteome Research

Streamlined Protocol for Deep Proteomic Profiling of FAC-sorted Cells and Its Application to Freshly Isolated Murine Immune Cells. - Abstract - Europe PMC

EPURISp: Combining Enzymatic Digestion, Ultrafiltration, and Rapid In Situ Sample Purification for High-performance Proteomics | Journal of Proteome Research

S-Trap, an Ultrafast Sample-Preparation Approach for Shotgun Proteomics | Journal of Proteome Research

SPIN enables high throughput species identification of archaeological bone by proteomics | Nature Communications

SCASP: A Simple and Robust SDS-Aided Sample Preparation Method for Proteomic Research - ScienceDirect
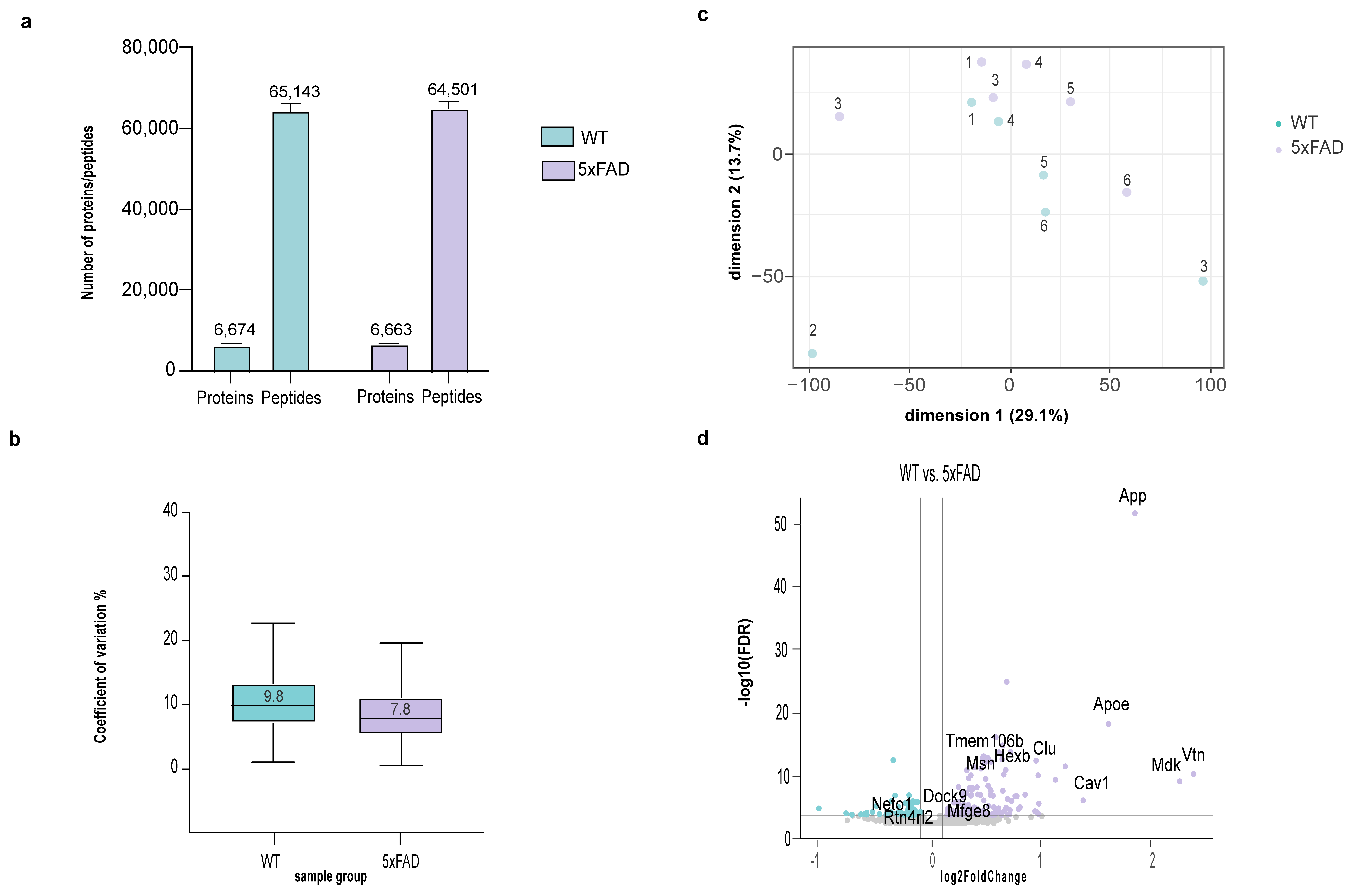
Cells | Free Full-Text | Suspension TRAPping Filter (sTRAP) Sample Preparation for Quantitative Proteomics in the Low µg Input Range Using a Plasmid DNA Micro-Spin Column: Analysis of the Hippocampus from the

Comparative proteomics analysis of in-gel digestion and sTRAP protocol... | Download Scientific Diagram
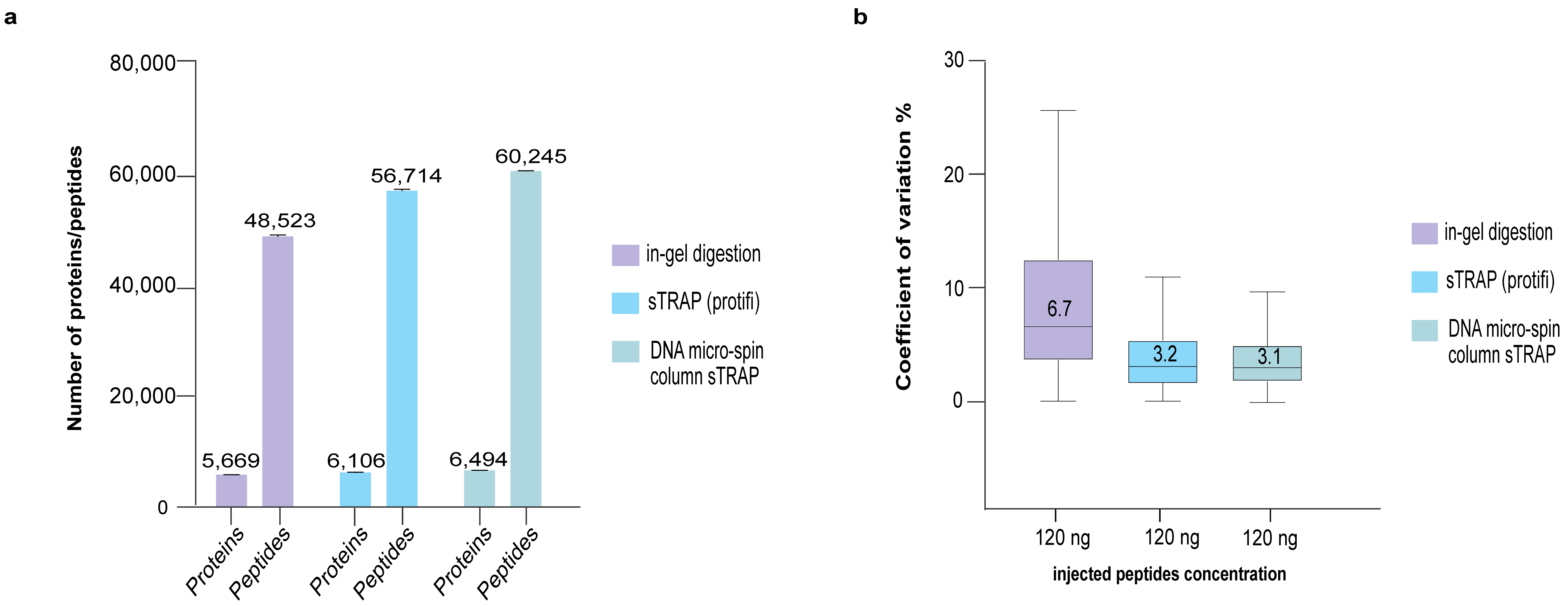
Cells | Free Full-Text | Suspension TRAPping Filter (sTRAP) Sample Preparation for Quantitative Proteomics in the Low µg Input Range Using a Plasmid DNA Micro-Spin Column: Analysis of the Hippocampus from the

Cells | Free Full-Text | Suspension TRAPping Filter (sTRAP) Sample Preparation for Quantitative Proteomics in the Low µg Input Range Using a Plasmid DNA Micro-Spin Column: Analysis of the Hippocampus from the
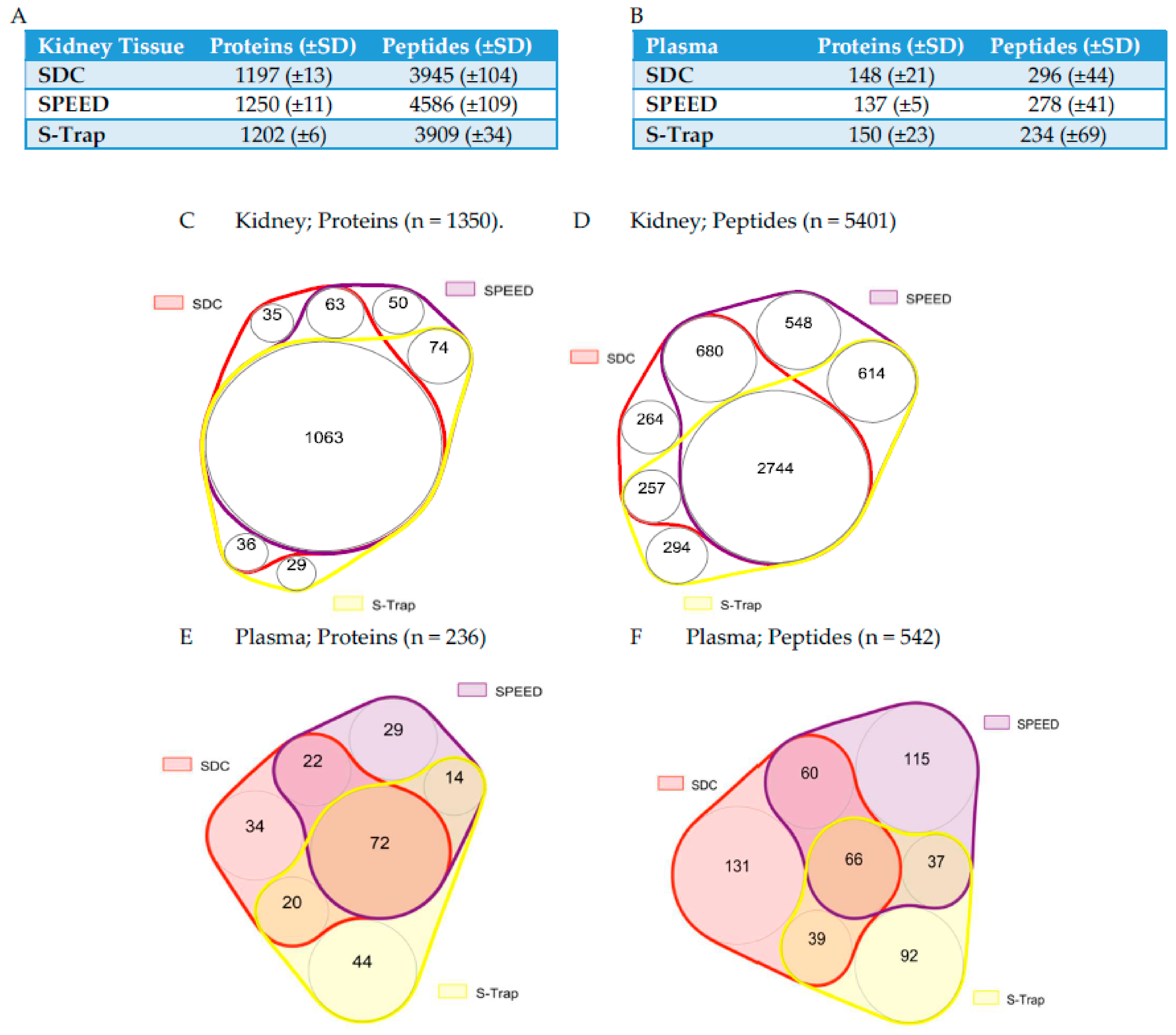
IJMS | Free Full-Text | Comparison of SPEED, S-Trap, and In-Solution-Based Sample Preparation Methods for Mass Spectrometry in Kidney Tissue and Plasma

Suspension trapping (STrap) sample preparation method for bottom‐up proteomics analysis - Zougman - 2014 - PROTEOMICS - Wiley Online Library